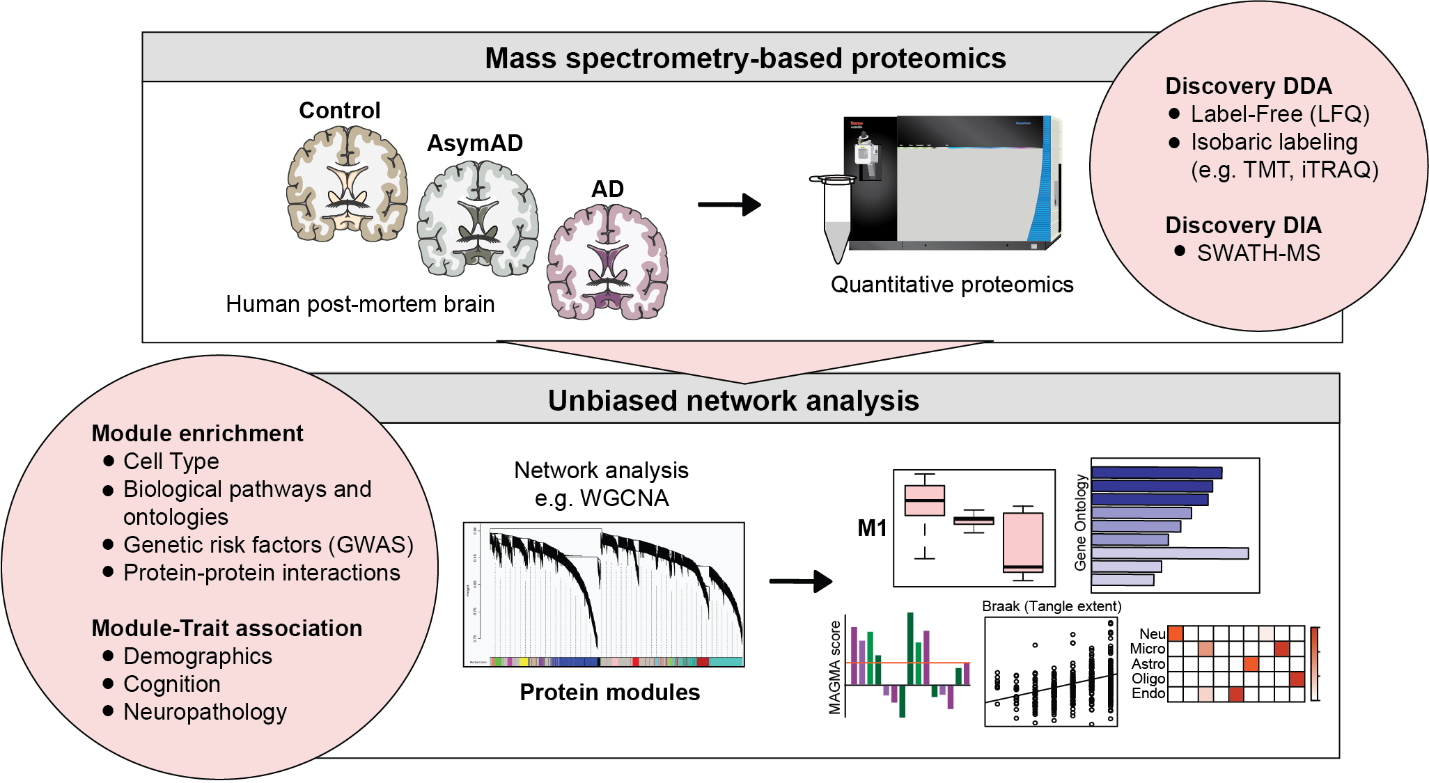
The expanding framework of brain disease pathophysiology has necessitated more holistic network-based “-omic” approaches to scientific investigation. The Seyfried Lab uses network proteomic analysis as a valuable tool for assessing pathophysiological changes in disease. This approach organizes complex proteomic data into unbiased groups or “modules” of protein co-expression that reflect various molecular, cellular, and circuit-level phenotypes.
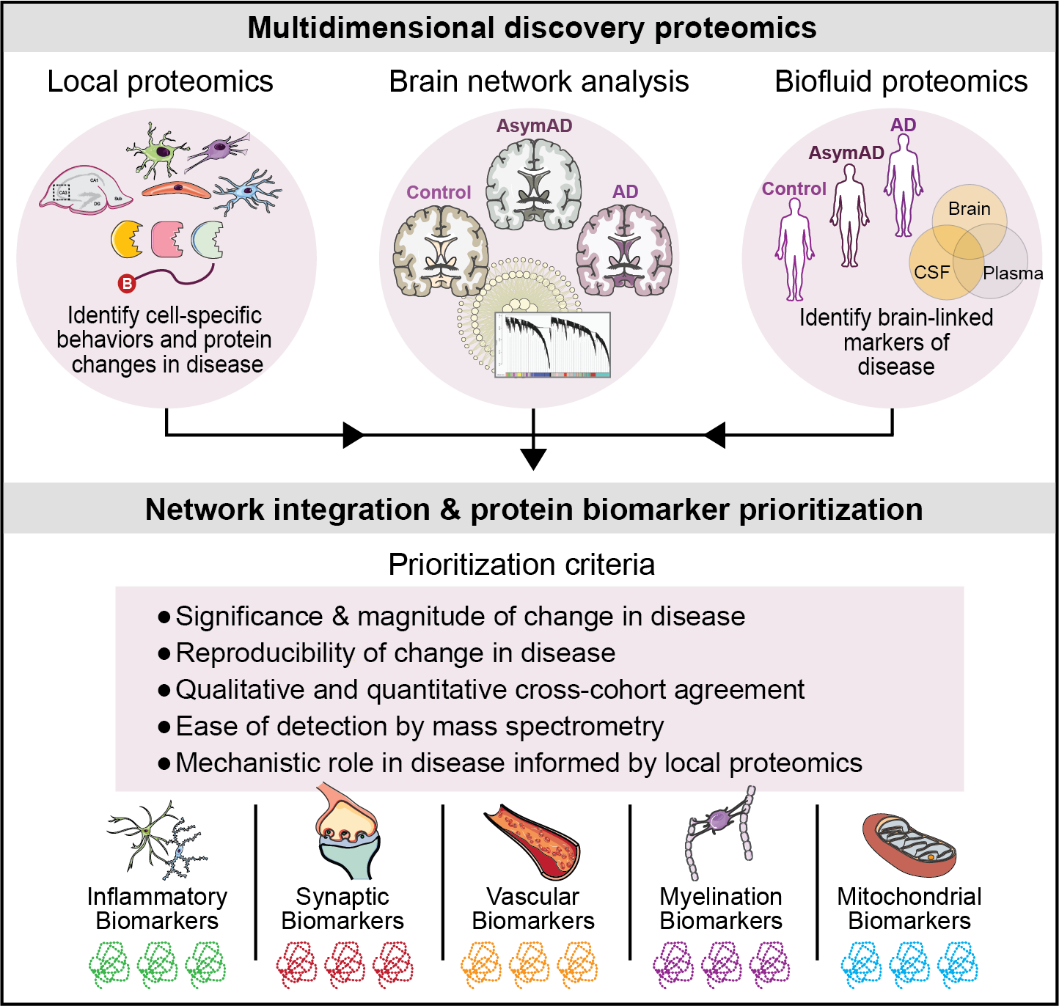
To increase the pathophysiological diversity among biofluid biomarkers of neurodegeneration, the Seyfried Lab has developed a novel approach that integrates the global brain network with proteomic analysis of biofluids. Moving forward, integrative analyses that successfully correlate proteomic signatures across brain, CSF, and plasma will prove key for the advancement of clinically relevant network-based biomarkers of disease.
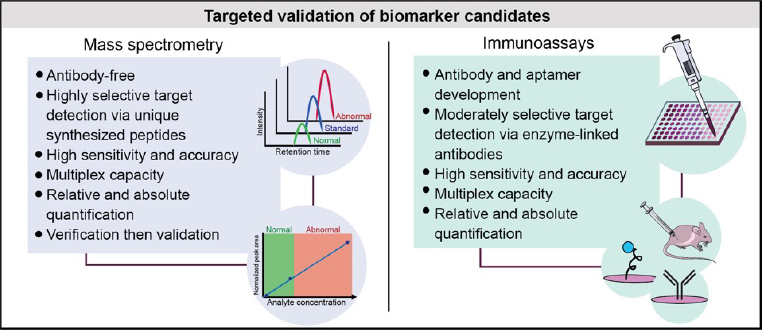
The current diagnostic framework for clinical AD and other neurological disorders fails to capture this biological diversity. The limited number of clinically established biomarkers stands to hinder advancements in diagnostic subtyping, disease monitoring, and therapeutic development. Approaching such a biologically heterogenous disease as a single entity in clinical trials may account for the many drug failures. A systems-based approach to biomarker discovery, as championed by the AMP-AD initiative, is growing in popularity among neurodegenerative disorders. The Seyfried Lab aims to translate the network AD brain proteome into systems-based, physiologically diverse, multiplex biomarker assays capable of advancing our clinical framework of disease.
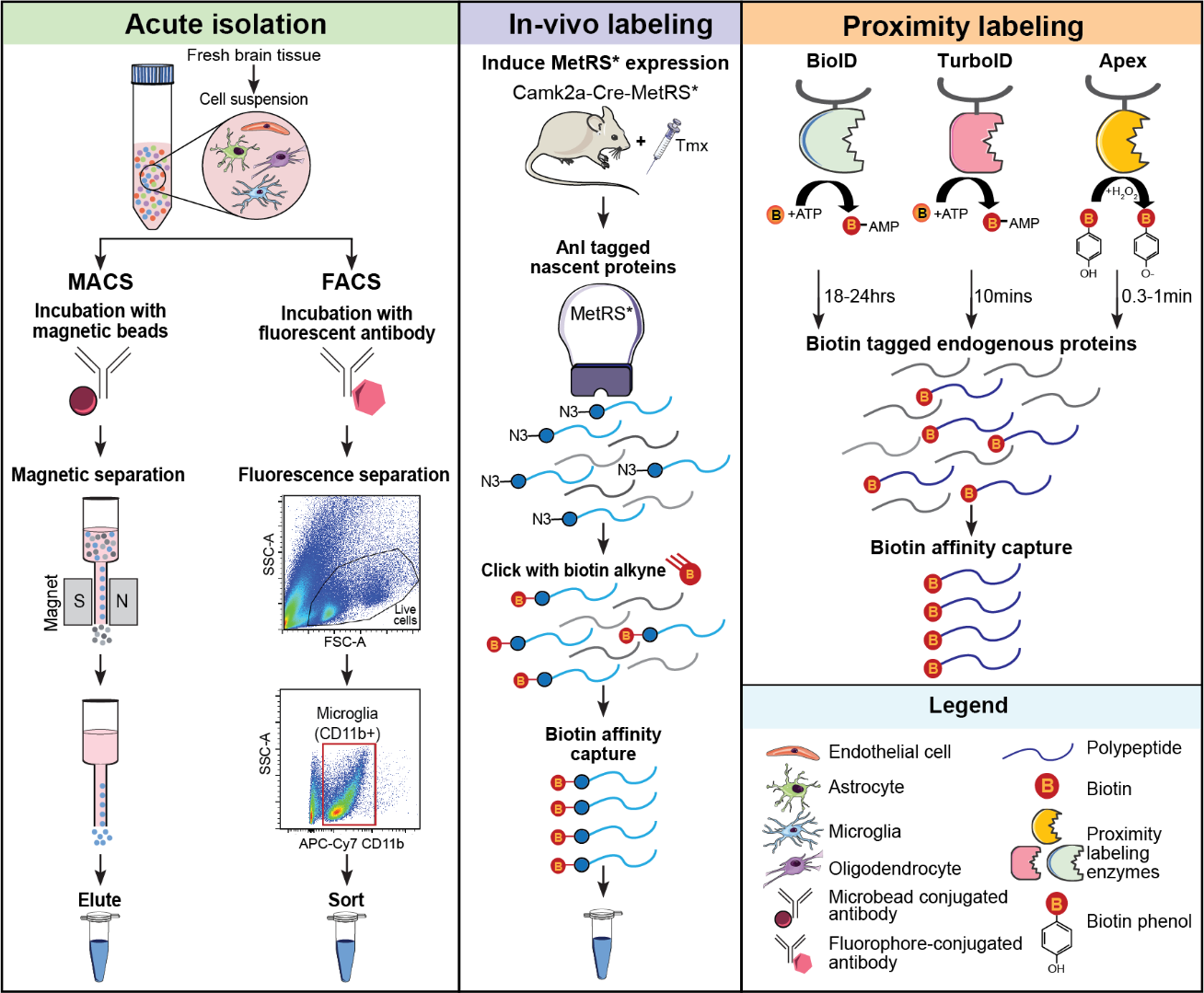
The next challenge for proteomic research is gaining the cellular and temporal resolution to further define the causative role of cell-mediated dysfunction in AD pathogenesis. Cell type-specific proteomics in the human brain is currently in its infancy, primarily due to the inability to isolate live, pure cell populations from frozen brain and limited access to fresh postmortem tissue. The Seyfried Lab aims to extend localized proteomic approaches to mouse models of AD pathology for confirming the network architecture and investigating causal molecular changes during the cellular phase of disease. Furthermore, cell type-specific proteomics in disease and aging mouse models can serve to de-convolute complex human brain data and provide cellular-level resolution to peripheral biomarkers. This work will significantly enhance our understanding of the aging-dependent course of cell type-specific perturbations, thus allowing us to pinpoint opportunities for therapeutic intervention and enhance biomarker development.
Dr. Nicholas Seyfried is also the Scientific Director of the Emory Integrated Proteomics Core (EIPC). The facility provides protein analytical services using innovative mass spectrometry (MS). Our major services include • Protein identification (in-gel or in-solution digestion) • Interactome analysis (on-bead digestion) • Post-translational modifications (PTM) • Isobaric (TMT) and Label-free quantification • Bioinformatic analysis support (additional charge may apply).